Cost-effective COVseq method could be used for large-scale genomic surveillance of VOCs
Published: 2021-07-01
The COVID-19 pandemic has not only changed societies and challenged healthcare, but also accelerated life science research. Over the last year and a half, a number of vaccines against COVID-19 have been developed in record time, and today large-scale vaccination campaigns are carried out in many countries worldwide. The open sharing of SARS-CoV-2 genome sequences has been key in facilitating the unprecedented rate of research. Research groups have made SARS-CoV-2 sequences available in, for example, European Nucleotide Archive (ENA) and/or GISAID. The large quantities of genomic data openly available to the research community has, in turn, enabled the study of SARS-CoV-2 spread and viral evolution, as well as the identification of novel SARS-CoV-2 variants. Throughout the current COVID-19 pandemic, a number of SARS-CoV-2 variants have been detected. The European Centre for Disease Prevention and Control (ECDC) designates some such variants as variants of concern (VOCs) and some others as variants of interest (VOIs). A variant designated as a VOC may, for example, have a higher transmissibility, cause a relatively increased severity of disease, and/or have greater immune escape capabilities (i.e. they are more able to infect those with previous immunity resulting from previous infection or vaccination). By contrast, VOIs may have specific genetic markers that are predicted to affect transmission, or be difficult to diagnose.
Genomic surveillance of SARS-CoV-2 could be used to identify new VOCs in a population, detect outbreaks of VOCs, and follow VOCs with greater immune escape abilities. In order to carry out effective SARS-CoV-2 genomic surveillance (using whole-genome sequencing (WGS) based on next-generation sequencing (NGS)), it may be necessary for a country/region to sequence thousands of samples each week. This means that cost-effective, reliable, and easily scalable methods for centralised sequencing are needed. Available commercial methods are both expensive and difficult to scale-up to larger quantities.
In a recently published study in Nature Communications, a collaboration of researchers at Karolinska Institute (Stockholm, Sweden), the ‘Amedeo di Savoia’ Hospital (Turin, Italy) and the Candiolo Cancer Institute (Turin, Italy) (first authors: Michele Simonetti, Ning Zhang, and Luuk Harbers, Corresponding author: Nicola Crosetto, Karolinska Institute) presented a cost-effective workflow called COVseq that can be used for surveillance of the viral SARS-CoV-2 genome. The method is both scalable and versatile for add-ons.
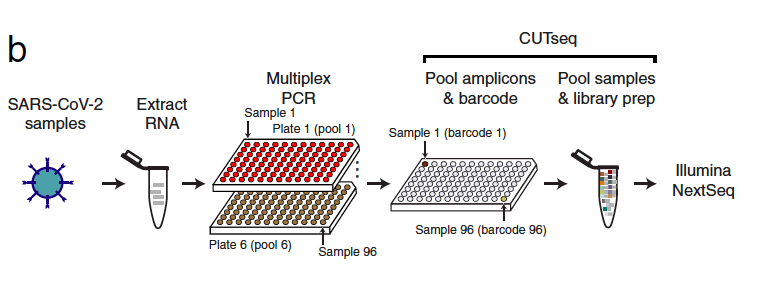
Simonetti, Zhang, and Harbers et al. proposed that COVseq can be used to prepare so-called multiplexed whole genome sequence (WGS) libraries. In short, the COVseq workflow include a multiplexed PCR assay where a large number of viral genome copies are created (originally developed by the U.S. CDC for the whole genome amplification of SARS-CoV-2) followed by CUTseq, a method that labels and then pools a high number of samples simultaneously. For the full step-by-step COVseq protocol see Simonetti, M. et al. COVseq is a cost-effective workflow for mass-scale SARS-CoV-2 genomic surveillance (Protocol Exchange, 2021).
The researchers prepared a single COVseq library (based on 29 SARS-CoV-2 positive RNA samples from patients from an Italian hospital). This was done in nanoliter volumes, in order to run the method at a low-cost. COVseq was found to have a similar ability compared to a standard commercial method used to identify small changes in the SARS-CoV-2 genome. The number of single-nucleotide variants (SNVs) per sample was highly correlated between COVseq and the commercial method NEBNext; 95.4% of the SNVs were detected by both methods. In addition, Simonetti, Zhang, and Harbers et al. also used COVseq to identify SNVs including those found in emerging VOCs by applying the COVseq to 245 additional SARS-CoV-2 positive samples.
In order to examine the cost-effectiveness of COVseq, the researchers performed a so-called real-life cost analysis based on a genomic surveillance program initiated for the Piemonte Region, Italy. This analysis showed that COVseq was highly cost-effective, with the cost per sample being less than 15 USD (incl. preparation of the library and sequencing).
In summary, the study shows that COVseq could be a useful tool for genomic surveillance of both SARS-CoV-2 and other viruses, such as influenza, even in low-income countries. ”The low per-sample cost enables COVseq to be used to sequence thousands of SARS-CoV-2 genomes per week and detect emerging VOCs at an early stage” says Michele Simonetti, a PhD student in the Crosetto lab.
Simonetti, Zhang, and Harbers et al. have shared the RNA-sequence data and deposited the BAM-files that were used to generate all plots in the European Nucleotide Archive (ENA) published under the project: PRJEB42601. The researchers have also made the code used for processing COVseq sequencing and custom MATLAB code used in the cost analysis (see Supplementary Notes) openly available in GitHub and Zenodo. All the GISAID data used in this study are described in Supplementary Data 7.
The research study was supported from the SciLifeLab National COVID-19 Research Program, financed by the Knut and Alice Wallenberg Foundation, the Swedish Research Council, as well as funds from the National Natural Science Foundation of China and the Chinese Postdoctoral Science Foundation and by private donations for COVID-19 research from Chiesi Pharma AB and Tetra Pak.
Data
- Full COVseq Protcol: Protocol Exchange, 2021
- RNA-sequence data and BAM files: PRJEB42601
- Code for processing COVseq sequences and cost analysis: GitHub repository and Zenodo
- Supplementary data related to the GISAID data used in the study: https://www.nature.com/articles/s41467-021-24078-9#MOESM9
Article
DOI: 10.1038/s41467-021-24078-9
Simonetti, M., Zhang, N., Harbers, L., Grazia Milia, M., Brossa, S., Huong Nguyen, T. T., Cerutti, F., Berrino, E., Sapino, A., Bienko, M., Sottile, A., Ghisetti, V., Crosetto, N. COVseq is a cost-effective workflow for mass-scale SARS-CoV-2 genomic surveillance. Nat Commun 12, 3903 (2021).