Interactions between fragments and SARS-CoV-2 nsp10 provide insight into the viral replications
Published: 2022-03-01
The COVID-19 vaccine program is the cornerstone of global pandemic management. Whilst vaccines were rolled out in record-time and have proven very effective, many important research questions remain. For example, there is still some uncertainty regarding the duration of vaccine protection, the optimal intervals for the administration of booster doses, the potential for vaccine escape by novel Variants of Concern (VoCs), among others. These questions, together with the fact that not everyone is able to be vaccinated, mean that the development of new drugs for treating SARS-CoV-2 infections is warranted. These drugs would act as a “second line defense” in the event of vaccine escape, and for individuals that do not respond well to vaccines, and are key for treating those that did not receive the vaccine for any reason.
Drugs to treat COVID-19 were also developed, and a limited number are now available for use. Research was conducted to explore repurposing drugs used for other purposes, in hopes that this would increase the numbers of drugs available relatively rapidly. This yielded some success. For example, both research and clinical studies showed that Dexamethasone decrease mortality among COVID-19 patients. However, the overall success of drug repurposing was limited.
Research has suggested that the non-structural protein 10 (nsp10) could prove a good target for drug therapy, due to its involvement in SARS CoV-2 replication. In particular studies have shown that the nsp10 forms a complex with viral nsps (nsp14 and nsp16) and has a vital role in RNA replication. Recently, the crystal structures of SARS-CoV-2 nsp10, both unbound and in different complexes have been reported (Rogstam and colleagues, 2020, see our data highlight).
In a recently published study, researchers from Lund University, MAX IV Laboratory, European Spallation Source ERIC, and University College London used fragment-based screening using nano differential scanning fluorometry and X-ray crystallography in order to identify ligands targeting SARS-CoV-2 nsp10 (first author: Frank Kozielski, corresponding authors: Wolfgang Knecht (Lund University) and Zoë Fisher (Lund University/European Spallation Source ERIC).
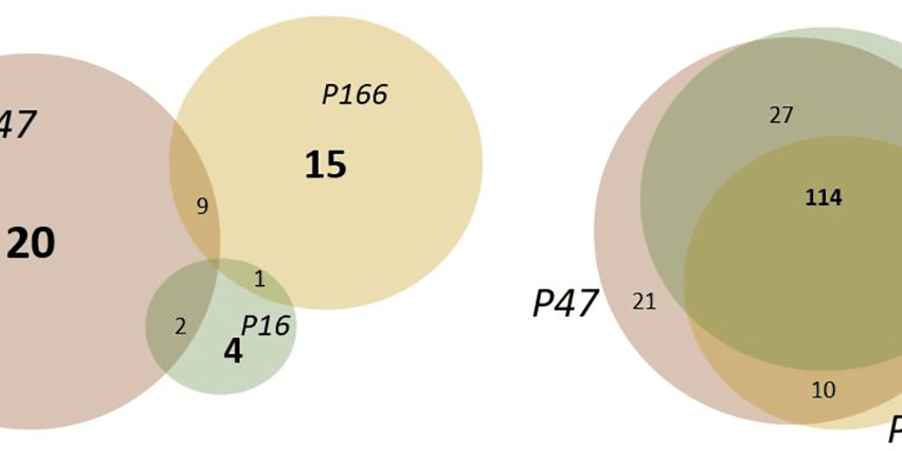
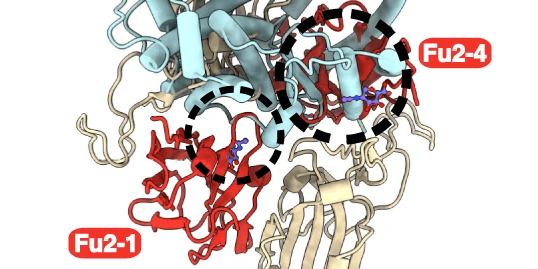
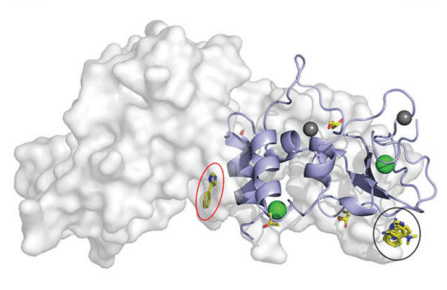
In order to identify fragments interacting with nsp10, Kozielski and colleagues employed two orthogonal assays in parallel; X-ray based fragment screening (XFS) and thermal shift assay (TSA). In brief, almost all available fragments from the FragMAX facility (107 of 110) were used in the XFS assay. After soaking of the crystals, diffraction data was collected using the BioMAX beamline of MAX IV. This resulted in four bound fragments from the FragMAX library. The identified fragments were located in two distinct sites. Using modelling, the researchers were able to show that one was located in the nsp14–nsp10 complex interface, and the other in the nsp16–nsp10 complex interface. The four found fragments were named Nsp10-VT00022, Nsp10-VT00221, Nsp10-VT00239, and Nsp10-VT00265. Whilst the Nsp10-VT00022 fragment could bind to two sites, as determined from the crystal structures, the other three bound to a single but identical site as the second VT-00022 location.
The two fragments that were found to stabilise nsp10 (VT00029 & VT00213) in the thermal shift assay (TSA) did not appear as hits in crystallographic screening. The researchers discussed this finding in detail, and concluded that the TSA assays give several outcomes; positive or negative Tm shifts, no shifts, or atypical curves. The assays performed in this study yielded a large number of atypical curves , and the results from the crystal assays could not be confirmed. The researchers thus surmised that the TSA is not suitable for selecting candidate fragments for co-crystallisability with nsp10, and moved forward with the XFS assay instead. The researchers also used Microscale thermophoresis (MST) experiments to quantify fragment affinities for nsp10. This showed that the interaction by nsp14 and 10 is weak , and therefore that complex formation could be disrupted by small molecules.
The structures from the study have been shared in the Protein Data Bank which in Europe is hosted by the European Bioinformatics Institute (EMBL EBI). The experimental data has been shared as a Zenodo entry.
“As a structural biologist keen to contribute to the global efforts in addressing the current and future pandemic(s), I am in strong support of sharing crystallographic data. Making data available as soon as possible will enable many scientists to work on various aspects: from understanding virus biology to drug & vaccine development. The PDB stands out as a central resource in this role - not only in hosting and curating the data, but also being actively involved in quality checking the deposited data. Our results are a small first step in the process of drug development against SARS-CoV-2, and we are hopeful that we can build on it to find solutions for the current and any future outbreaks,” says Zoe Fisher.
In summary, Kozielski and colleagues are the first to have successfully identified and characterised the interactions between fragments and SARS-CoV-2 nsp10. They have also contributed important information about the protein–protein complexes involving nsp10 in the viral replication machinery, which may be useful for future studies. The researchers now plan to continue this research. They will use the data from these fragments as a starting point to develop more potent analogues, using fragment growing techniques and structure-based drug design.
Data
- Protein Data Bank 7ORR: Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00022
- Protein Data Bank 7ORU: Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00221
- Protein Data Bank 7ORV: Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00239
- Protein Data Bank 7ORW: Non-structural protein 10 (nsp10) from SARS CoV-2 in complex with fragment VT00265
- Zenodo entry: Repository containing experimental data from a crystallographic fragment screening campaign conducted against the Non-structural protein 10 (nsp10) from SARS-CoV-2 at the FragMAX facility at MAX IV Laboratory.
Article
DOI: 10.1039/D1CB00135C
Kozielski, F., Sele, C., Talibov, V.O., Lou, J., Dong, D., Wang, Q., Shi, X., Nyblom, M., Rogstam,A., Krojer, T., Fisher, Z., and Knecht. W. Identification of fragments binding to SARS-CoV-2 nsp10 reveals ligand-binding sites in conserved interfaces between nsp10 and nsp14/nsp16 RSC Chemical Biology, 1 (1) (2022).
Funding
This work was supported by the Swedish Research Council, the Swedish Governmental Agency for Innovation Systems (Vinnova), and the Swedish Research Council for Sustainable Developmen (Formas). The project was funded by ESS for projects related to COVID-19, and it profited from Lund University’s and its faculties’ infrastructure investments as support of the Lund Protein Production Platform (LP3).