Interferon signatures in SARS-CoV-2 infected liver cells could provide knowledge for new antiviral therapies
Published: 2021-07-06
The research community has responded rapidly to the current COVID-19 pandemic, which has caused a global health crisis. The SARS CoV-2 virus has been shown to have similar clinical features to previously well-known coronaviruses such as SARS-CoV, and MERS-CoV. In addition, research has indicated that more severe forms of coronavirus diseases are associated with dysregulation of the human type-I interferon (IFN-I) response, but further studies into this topic are warranted. Previous studies (e.g. McNab (2005)) have found that Type I IFNs have diverse effects on innate and adaptive immune cells during different infections (caused by viruses, bacteria, parasites or fungi). Type I IFNs have been found to be important for host defence against viruses, but more recent studies indicate that it can also cause pathogenesis in acute viral infections, e.g. influenza.
The interferon-response has been shown to be a key feature of COVID-19 infection. Previous studies have shown that Type-I interferons (IFN-I), primarily IFN-α and IFN-β, are part of the earliest antiviral innate immune responses following the viral SARS-CoV-2 infection, and also play an important role in pathogenesis.
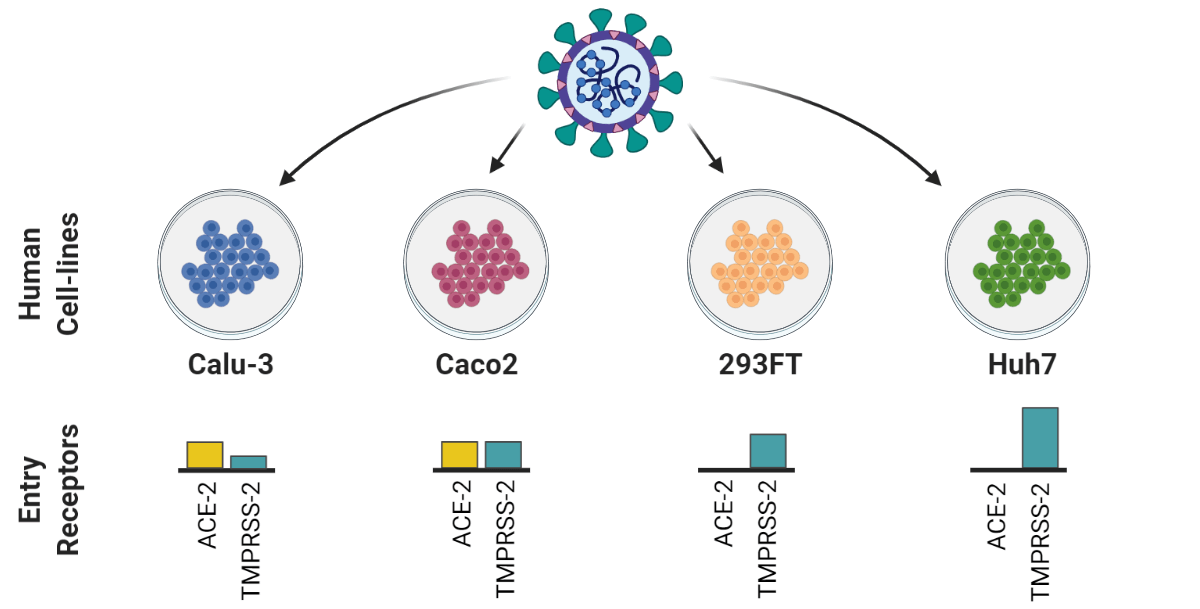
A recently published article in Nature publishing group’s Cell Death Discovery, carried out as a collaboration between researchers from the Karolinska Institute, Umeå University and Public Health Agency Sweden, (First authors: Xi Chen and Elisa Saccon. Corresponding author: Assistant Professor Soham Gupta, Karolinska Institute), studied the Type-I IFN response in SARS-CoV-2 infected Huh7 cells.
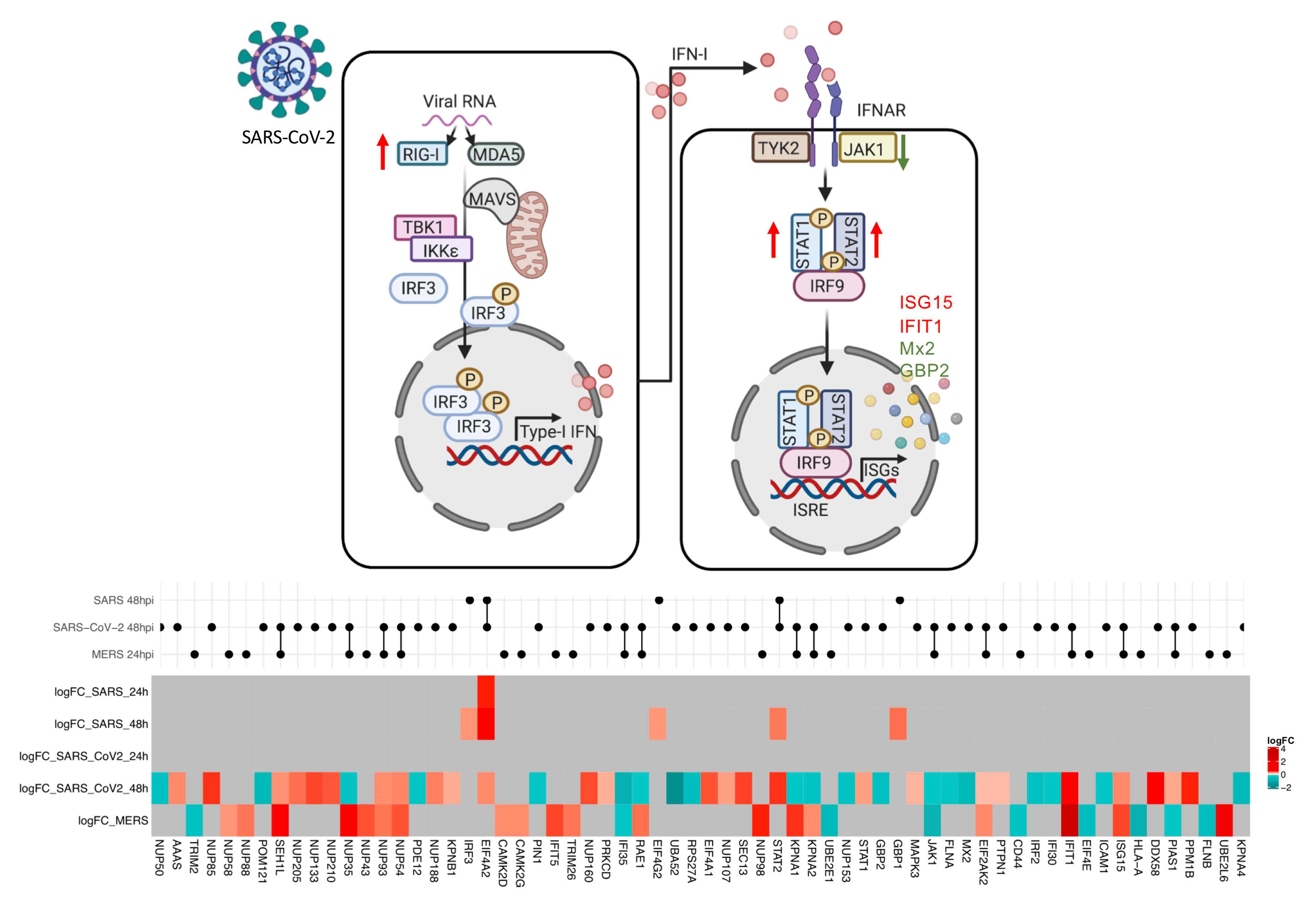
In the study, Chen and Saccon et al. used a proteomics-based approach to show that SARS-CoV-2 infection can induce a delayed and dysregulated IFN-I signaling in Huh7 cells (human liver cell line). In addition, the results showed SARS-CoV-2 virus could inhibit retinoic acid-inducible gene I (RIG-I) mediated IFN-β production and that IFN-I pretreatment could reduce the susceptibility of Huh7 cells to SARS-CoV-2, though this was not found for post-treatment. The results also showed that senescent (older, less physiologically useful) Huh7 cells, despite showing an accentuated IFN-I response, were more susceptible to be infected by SARS-CoV-2, and the virus effectively inhibited the interferon-induced protein IFIT1 in the senescent cells.
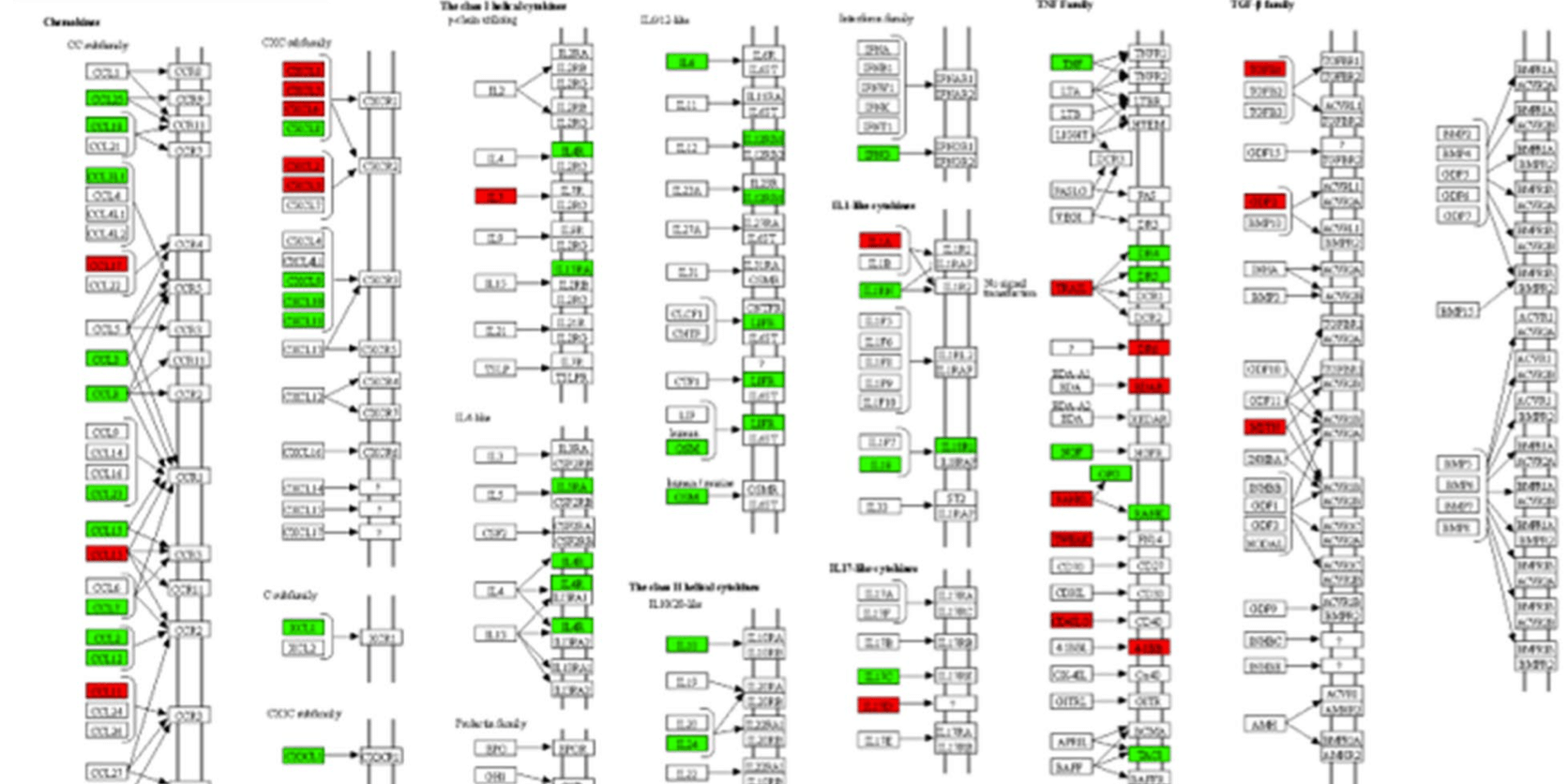
They also measured the global proteomic changes using quantitative proteomics to investigate the differences in pathogenicity between the new SARS-CoV-2 and two well-studied members of the coronavirus family (SARS-CoV and MERS-CoV) using Huh7 cell assays. The analysis focused on the IFN-response, specifically on the proteins that were found to be differentially regulated in any of the viruses investigated. The results revealed some overlap between SARS-CoV-2 and MERS-CoV. Briefly, the regulation of 13 proteins was found to be affected by both SARS-CoV-2 and MERS-CoV. Of these, eight were found to have their regulation affected in the same way (six (ISG15, IFIT1, EIF2AK, NUP54, NUP93, and SEH1L) were upregulated, two (JAK1 and IFI35) were downregulated), and the remainder had their regulation affected differently (PIAS1 was upregulated by SARS-CoV-2 and downregulated by MERS-CoV, while nuclear receptors (e.g. KPNA1, KPNA2, and RAE1) were downregulatd by SARS-CoV-2 and upregulated by MERS-CoV). The researchers concluded that there is a distinct, differential regulatory signature of interferon-related proteins between SARS-CoV-2, SARS-CoV, and MERS-CoV. While the results provided a perspective of immune regulation by coronaviruses, the results may also indicate that therapeutic strategies based on observations from SARS-CoV and MERS-CoV viruses could have to be used with caution.
“Interferon response is one of the first lines of antiviral defence, and plays an important role in shaping the disease course of SARS-CoV-2 infection. An early robust interferon response can suppress the virus, and has been correlated with a positive disease outcome. However, in the later stages of the disease, SARS-CoV-2 causes dysregulated interferon signaling. This is what has been observed in our in vitro Huh7 cell infection model - that the virus can inhibit early interferon response, cause dysregulated interferon signaling in the later stages of infection, and that interferon treatment prior to infection can suppress the virus, although interferon treatment following infection has no inhibitory effect on the virus. This tells us that, for any interferon-based treatment, the timing is critical and that such treatment should be used with caution. The interferon signaling stimulates a wide range of genes (known as interferon stimulatory genes (ISGs)) with antiviral properties and they could be harnessed as novel therapeutics against viruses. Here we have mapped the ISGs induced or suppressed by SARS-CoV-2, MERS-CoV and SARS-CoV-1 infection in Huh7 cells. However, there could be cell-type specific differences. We are continuing to study the protective role of different ISGs against several human pathogenic viruses.” says the corresponding author of the article, Assistant Professor Soham Gupta.
In summary, Chen and Saccon et al. propose that the recent findings can contribute to providing a better understanding of how the SARS-CoV-2 virus can affect and regulate the human cellular interferon response. The results highlight the importance of further studies focused on interferon-stimulated genes as well as studies to investigate the role interferon-stimulated genes play in inhibition of SARS-CoV-2 pathogenesis. Increased understanding of the interferon response is an important step in developing novel antiviral strategies, both for COVID-19 and future pandemics.
The researchers have shared the raw mass spectrometry in ProteomeXchange Consortium via the PRIDE partner repository (dataset identifier: PXD023450). Additionally, the researchers have deposited all the bioinformatic analysis codes (for example, for proteomics, transcriptomics data and heatmaps) in GitHub. Additional datasets generated as part of this study may be requested from the corresponding author.
The researchers within this study were supported by grants from Swedish Research Council, Karolinska Institute Stiftelser och Fonder, Åke Wibergs Stiftelse, Innovative Medicines Initiative 2 Joint Undertaking, European Union’s Horizon 2020 and Swedish Cancer Society. Open access funding provided by Karolinska Institute.
Data
- Raw mass spectrometry proteomics data: ProteomeXchange Consortium, accession number PXD023450
- Code for analysis and visualisations: GitHub
- Other datasets available upon request from corresponding author (Assistant Professor Soham Gupta)
Article
DOI: 10.1038/s41420-021-00487-z
Chen, X., Saccon, E., Appelberg, K. S., Mikaeloff, F., Esneider Rodriguez, J., Sá Vinhas, B., Frisan, T., Végvári, Á., Mirazimi, A., Neogi U., Gupta S. Type-I interferon signatures in SARS-CoV-2 infected Huh7 cells. Cell Death Discovery 7 (114) (2021)