Nanobodies promising new therapeutics for treatment of SARS CoV-2
Published: 2021-02-16
The COVID-19 pandemic has strained the healthcare systems and posed challenges to economic and social activity in many countries. Currently, a number of vaccines have been rolled out in record time. However, vaccines may not be suitable for all patients. Therefore, additional therapeutics or treatments of high-risk patients or patients already infected with the SARS CoV-2 virus are warranted.
Previous research has proposed neutralizing antibodies or related molecules as potential immediate and direct-acting antiviral agents (see also our data highlight about synthetic nanobodies). Camelid heavy chain-only antibodies (VHHs), nanobodies, can offer a cheap, easy way to produce antiviral agents for passive immunization. Nanobodies have favourable biochemical properties, i.e., high thermostability and deep tissue penetration. In addition, due to their small size nanobodies can bind to the SARS CoV-2 virus more efficiently.
A recently published study which was a result of collaboration between Swedish, German, and American researchers (first author: Paul-Albert Koenig, corresponding authors: B. Martin Hällberg, Nicholas C. Wu, and Florian I. Schmidt) characterized the mechanisms by which nanobodies neutralize or prevent of infection in molecular detail.
Koenig and colleagues first immunized one alpaca and one llama with the receptor binding domains (RBD) of SARS-CoV-2 spike as well as formalin-inactivated SARS-CoV-2 to elicit neutralizing heavy chain-only antibodies. Four novel neutralizing nanobodies (VHH E derived from the llama, and VHHs U, V, and W from the alpaca) that target the RBD of SARS-CoV-2 spike were identified. In addition, Koenig and colleagues demonstrated the inhibitory effect of the nanobodies, preventing the virus from spread, using different in vitro human cell models.
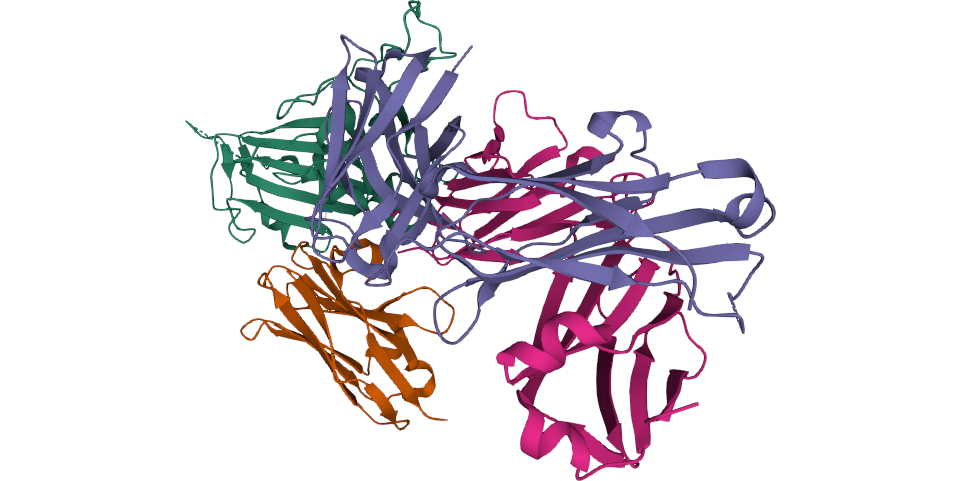
In the next stage, the researchers used X-ray crystallography and electron cryomicroscopy (cryo-EM) to study how the different nanobodies bind to the spike protein on the SARS CoV-2 virus and defined two distinct binding epitopes. The VHH E and U, V, and W bind to distinct epitopes on the RBD. The results showed that VHH E employ its complementarity determining region (CDR) 1 and its unusually long, 22-amino acid CDR3 to bind the ACE2 binding site on the RBD.
A biparatopic construct (VE) of VHH V and VHH E connected via a linker – a design guided by cryo-EM performed at the Karolinska Institutet’s 3D-EM facility – showed more than 100-fold improved neutralizing activity. In addition, the VE construct was found to eliminate the emergence of escape mutants.
In summary, the use of structure-based multivalent nanobodies may have potential clinical applications owing to increased neutralizing activity and protect from rapid emergence of novel mutant strains of SARS CoV-2. This research shows that nanobodies may be a promising way to develop additional new therapeutics for COVID-19. The company Dioscure SE has licensed the rights to develop therapeutics based on the research and aims to perform clinical trials during the spring of 2021.
This project has received funding from a number of Swedish, German and American funders, i.e., Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) the Emmy Noether Programme and Germany’s Excellence Strategy The Klaus Tschira Boost Fund, Bundesministerium für Bildung und Forschung (BMBF), European Union’s Horizon 2020 research and innovation program (CoroNAb consortium), Swedish Research Council, Knut and Alice Wallenberg Foundation, Bill and Melinda Gates Foundation. Cryo-EM data has been collected at the Karolinska Instititute 3D-EM Facility. A number of pending patents have been files by the authors.
Article
Koenig, P-A., Das, H., Liu, H., Kümmerer, B. M., Gohr, F. N., Jenster, L-M., Schiffelers, L. D. J., Tesfamariam, Y. M., Uchima, M., Wuerth, J. D., Gatterdam, K., Ruetalo N., Christensen, M. H., Fandrey, C. I., Normann, S., Tödtmann, J. M. P., Pritzl, S., Hanke, L., Boos, J., Yuan, M., Zhu, X., Schmid-Burgk, J. L., Kato, H., Schindler, M., Wilson., I. A., Geyer, M., Ludwig, K. U., Hällberg, B. M., Nicholas C. Wu, N. C., & Schmidt, F. I. Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape. Science, 371 (6530) (2021).
Data
Crystallographic coordinates and data have been deposited in the Protein Data Bank under accession codes 7KN5 (RBD + VHH E + VHH U), 7KN6 (RBD + VHH V + CC12.3), and 7KN7 (RBD + VHH W + CC12.3). The cryo-EM density maps have been deposited in the Electron Microscopy Data Bank, under accession codes EMD-23018 (Spike + VHH E), EMD-11978 (RBD + VHH E; will be released publicly shortly), EMD-11977 (Spike + VHH V; will be released publicly shortly), EMD-11981 (Spike + VHH VE; will be released publicly shortly), and EMD-11980 (RBD + VHH VE). In addition, the atomic coordinates have been deposited in the Protein Data Bank, under accession codes 7KSG (Spike + VHH E), 7B14 (RBD + VHH E; will be released publicly shortly), 7B11 (Spike + VHH V; will be released publicly shortly), 7B18 (Spike + VHH VE; will be released publicly shortly), and 7B17 (RBD + VHH VE).