Direct RT-PCR used to monitor Omicron BA.1/BA.2 variant transition in Sweden (Update)
Published: 2022-08-15
Update: This data highlight has been updated upon formal publication of the article. It was originally featured on the portal in April 2022 as a preprint article.
Since the start of the COVID-19 pandemic, multiple SARS-CoV-2 variants have emerged in different parts of the world. The European Centre for Disease Control (EDCD) classified several of them as Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), Delta ( B.617.2), and Omicron (BA.1 and B.1.1.529) as variants of concern (VOC). The three major waves that have swept across the world have been associated with different VOCs; with Alpha causing the wave in early 2020, Delta in mid 2021, and Omicron from late 2021 to early 2022. Omicron was first detected in South Africa and Botswana in November 2021. WHO declared it to be a VOC later that same month and, by early December 2021, it had already spread to over 50 countries. By mid-January 2022, over 50 million infections were detected worldwide daily.
In Sweden, Omicron BA.1 had already become widespread when a sub-variant of Omicron (B.1.1.529.2, or BA.2) was first detected in the country. Both Omicron variants have since been spreading in Sweden. Research efforts have occurred at unprecedented levels throughout the COVID-19 pandemic, especially in relation to VOCs and vaccines, but further research on the Omicron variant is clearly needed to tackle this latest VOC.
In a recent pre-print researchers from Karolinska Institutet, Department of Medical Biochemistry and Biophysics, (First author: Antonio Lentini, Corresponding author: Björn Reinius) used a custom variant-typing RT-PCR assay to track the shift in dominance from Omicron BA.1 to the BA.2 Omicron sub-variant in the Swedish population in early 2022.
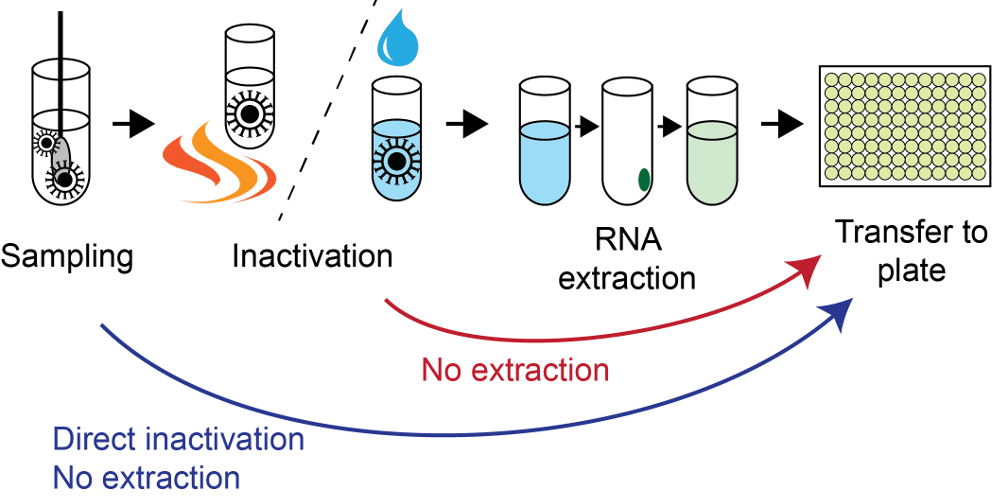
The SARS-CoV-2 RT-PCR assay developed by the US Center for Disease Control (CDC) (Lu et al. 2020), was originally given emergency authorization by the FDA in Feb 2020. Lentini and colleagues developed a modified version of the CDC RT-PCR assay, simultaneously detecting general SARS-CoV-2 infection status (N1-FAM probe), human RNaseP sample integrity (RP-HEX probe) and Omicron BA.1-variant status (SBA1-Cy5 probe), leveraging BA.1-specific mutations in the spike (S) gene. The direct RT-PCR assay represents an improved, multiplex version of the RNA-extraction-free protocol described by the Reinius lab previously Smyrlaki et al. 2020. A previous data highlight also describes this study.
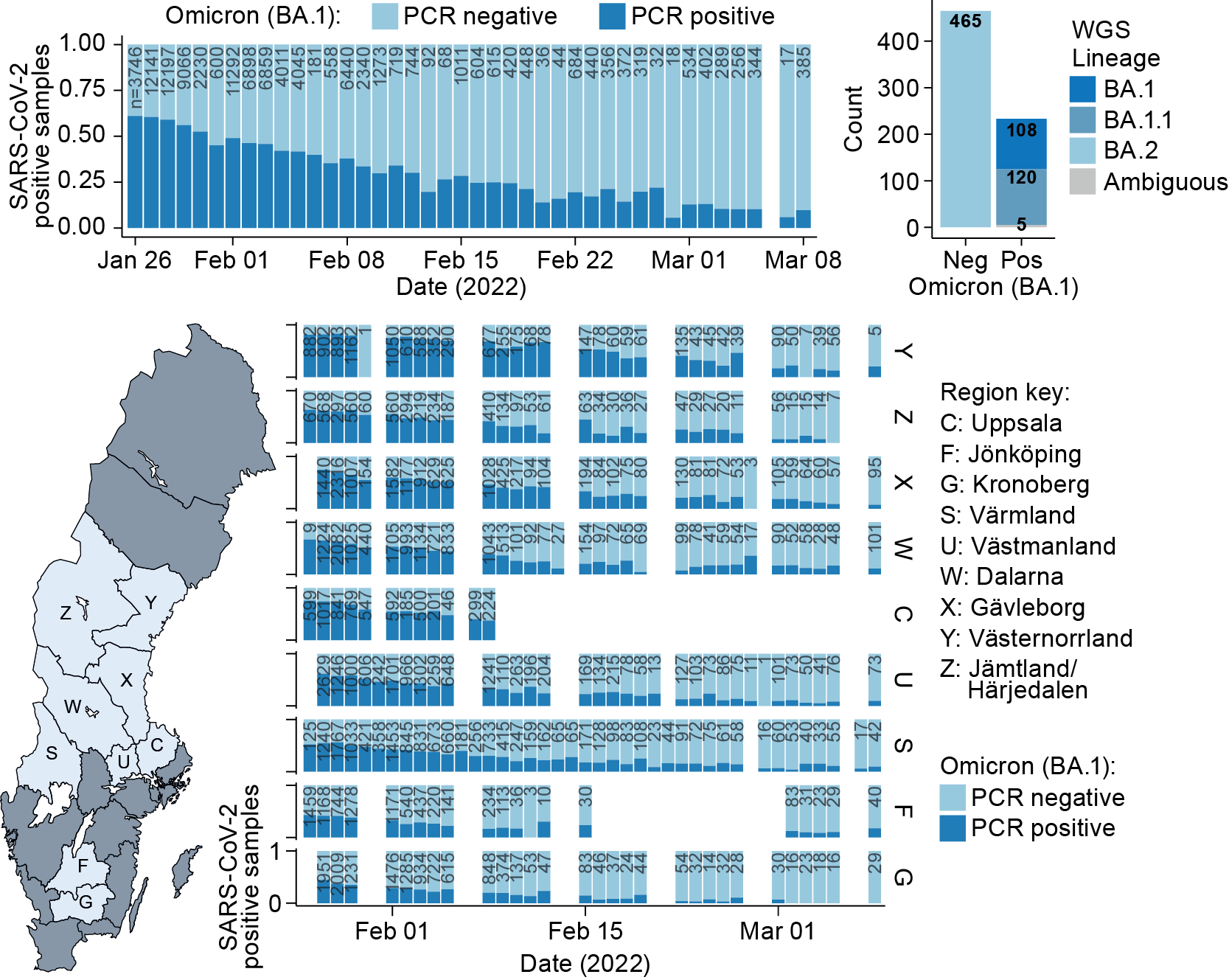
In brief, the new direct RT-PCR assay handles greater sample and reaction volumes, has increased sensitivity, and reports general SARS-CoV-2 infection status and Omicron BA.1 variant status simultaneously in a single RT-PCR reaction. Validation was completed by performing genotyping using the new RT-PCR assay and Thermo Fisher TaqMan SARS-CoV-2 Mutation Panel Assay in parallel. Test samples that were already confirmed to be Omicron-BA.1 by whole-genome sequencing (WGS) (N=133) were used during validation, and 100% consistency was found between the two tests.
After validating the direct RT-PCR assay, the assay was used to monitor the Omicron BA.1/BA.2 wave across central Sweden day-by-day between 26th January – 8th March, 2022. A total of 174,933 nasopharyngeal samples were analysed with the assay, with genome sequencing of 801 samples demonstrated >99% agreement with BA.1 calls obtained directly in the RT-PCR. Further, while performing WGS of Omicron BA.2, the researchers observed the BA.2-specific ORF3a:H78Y mutation. Denmark was one of the first countries to be swept by the Omicron-wave, and this specific BA.2 mutation was first detected in Denmark. These results suggest a common Danish/Swedish origin of this specific mutation, and this warrants further research. The results show in real-time how the Omicron BA.1 variant was outcompeted by BA.2 during the latest wave.
Whilst the first COVID-positive nasopharyngeal samples in January 2022 were found to be dominated by Omicron BA.1, by March almost all positive samples resulted from the more transmittable BA.2 sub-variant. Importantly, in cases resulting from Omicron BA.2, the viral RNA levels detected in the nose/throat samples were found to be two-fold higher than for Omicron BA.1. An increased viral load in the upper pharynx upon BA.2 infection could provide part of the explanation why BA.2 is more transmissible, and able to outcompete the BA.1 variant.
For more information, also see the news article on Karolinska Institutet’s page.
Data
- Computational code is available here.
- Assembled SARS-CoV-2 genomes have been deposited to GISAID (EPI_ISL_11414777-11415389 and EPI_ISL_11450500-11450677).
- Raw sequencing data has been deposited to ArrayExpress (accession: E_MTAB-11650).
Article
Lentini, A., Pereira, A., Winqvist, O., Reinius, B (2022). Monitoring of the SARS-CoV-2 Omicron BA.1/BA.2 lineage transition in the Swedish population reveals increased viral RNA levels in BA.2 cases. Med. https://doi.org/10.1016/j.medj.2022.07.007.
Funding
This research was supported by the SciLifeLab/KAW national COVID-19 research program project grant, the Swedish Research Council and the Ragnar Söderberg Foundation