New phenomics method for antiviral drug discovery may help to fight COVID-19
Published: 2021-08-02
The current pandemic has increased the demand for rapid development of COVID-19 treatments, therefore increasing the need to develop methods to screen for novel drugs and drugs that could be repurposed. SARS-CoV-2 belongs to the RNA family of viruses, a group of viruses known for their fast mutation rates and resilience to antiviral drugs. Therefore, investigations into how the viruses affect host cells are needed in order to discover novel drug targets.
Phenomics (image-based morphological profiling of cells) can be used to study both biological and chemical perturbations. An advantage of using a phenomics approach to study cellular responses to virus infection is the opportunity to combine high-content imaging and multiparametric analysis of the host cells.
A recently published article in BMC Biology (accepted July 8th, published online August 2nd 2021) (First author Jonne Rietdijk, PIs: Ola Spjuth Department of Pharmaceutical Biosciences and Science for Life Laboratory, Uppsala University, and Marjo-Riitta Puumalainen, Karolinska Institute. Corresponding author: Jordi Carreras-Puigvert at Department of Pharmaceutical Biosciences and Science for Life Laboratory, Uppsala University) proposes a phenomics approach for antiviral drug discovery.
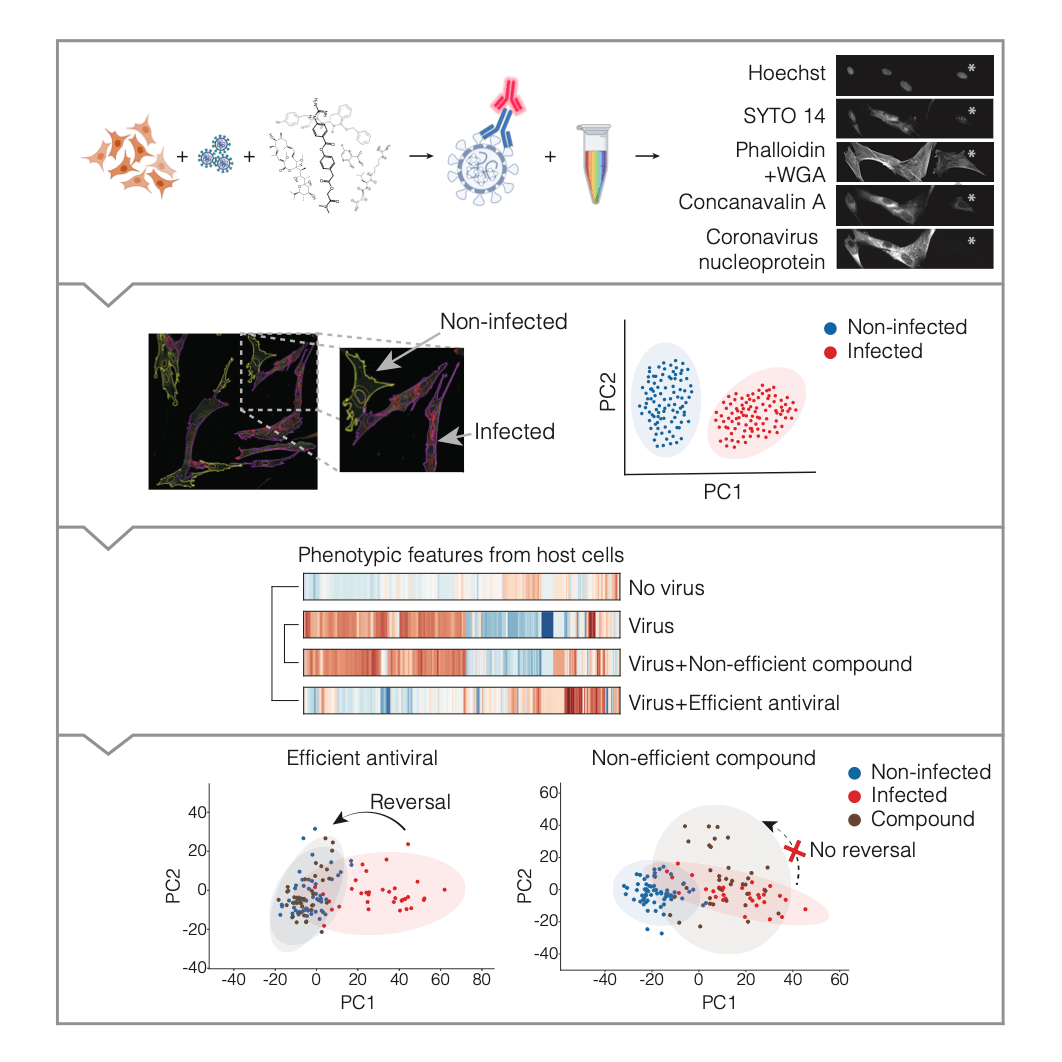
Rietdijk and colleagues investigated how viral infection affects the host cells using a method for untargeted phenotypic drug screening of virus-infected cells. The researchers combined a modified Cell Painting assay with antibody-based detection of viral infection in a single assay. They designed an automated image analysis pipeline for segmentation and classification of virus-infected and non-infected cells, followed by extraction of morphological properties. The Cell Painting assay is used for morphological profiling. The assay uses six multiplexed fluorescent dyes that are imaged in five channels and reveal eight cellular compartments involved in different biological pathways. The resulting images can be used to study, for example, compound toxicity, mechanism of action, and morphological disease signatures.
In this study, Rietdijk and colleagues combined Cell Painting with a virus nucleoprotein antibody staining as a tool to profile or screen for antiviral drugs. This made it possible for them to capture in-depth morphological profiles of both virus infected and drug-treated host cells. This proposed novel phenomics approach includes several steps: viral infection, compound treatment, a cytochemistry protocol, an image analysis pipeline, and then data analysis and visualisation.
In addition, the researchers also demonstrated the proposed phenomics method using a compound panel of known and novel antivirals on MRC5 human lung fibroblasts infected with Human coronavirus 229E (CoV-229E). The researchers were able to detect a distinct virus-induced phenotype for MRC-5 cells infected with CoV-229E. Notably, the results showed that the novel phenomics approach could identify antiviral compounds, i.e., Remdesivir, E-64d, TH3289, TH6744 and TH5487, that could reverse the virus-induced morphological profile towards a healthy state. For an overview and introduction to the phenomics method please see the following short YouTube video.
In summary, the results from the study suggest that the novel phenomics approach can provide researchers with valuable information about the effect of both viruses and drugs on host cells. Development of novel methods is an important part of COVID-19-related drug discovery. Rietdijk and colleagues highlight that the novel phenomics approach may be useful for morphological profiling of novel antiviral compounds on both infected and non-infected cells.
In order to share data as early as possible, the article was made public on the BioRxiv preprint server on Jan 13th 2021.
The researchers have publicly shared all image data, the image analysis pipelines (quality control, illumination correction, and feature extraction), extracted features, as well as an example analysis pipeline. These are available in the SciLifeLab Data Repository (hosted by FigShare). All data generated or analysed during this study were also included in the supplementary information made available alongside the published article (see licence agreement). In addition, data analysis code is available in Github.
The study was supported by SciLifeLab National COVID-19 Research Program and Knut and Alice Wallenbergs stiftelse (KAW 2020.0182). In addition, the project also received funding from the Swedish Research Council, FORMAS and the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement no 722729 and SSF.
For more information about the Pharmaceutical Bioinformatics group activities please visit pharmb.io.
Data
- Short video providing an overview and introduction to the phenomics method: YouTube
- All image data, the image analysis pipelines (quality control, illumination correction and feature extraction) and extracted features: Figshare
- Representative images of compounds and assays, information on analyses, statistical analyses results and numerical data related to figures: Supplementary Information
- Code for data analyses: GitHub
Article
DOI: 10.1186/s12915-021-01086-1
Rietdijk, J., Tampere, M., Pettke, A., Georgiev, P., Lapins, M., Warpman-Berglund, U., Spjuth, O., Puumalainen, M-. R., Carreras-Puigvert, J. A phenomics approach for antiviral drug discovery. BMC Biology 19 156 (2021)