On this page we have compiled current knowledge on Omicron, the latest SARS-CoV-2 variant to be designated as a Variant of Concern (VOC). This includes available data, ongoing research efforts, initial findings, among other things related to Omicron. Please see this page for information on other VOCs.
We make all possible efforts to track preprints, publications, and other resources related to Omicron and link them below. Researchers and organisations from around Sweden are also welcome to submit their datasets and to make relevant announcements on this page. If you feel like something is missing that should be included, or you wish to make a submission/announcement, please use the forms below or email us at datacentre@scilifelab.se. The team behind the COVID-19 Data Portal continuously monitors incoming suggestions, and new items are published as soon as possible.
Available data
This section presents a list of the available data related to Omicron (or related work that could benefit research into this variant) shared by researchers affiliated to a Swedish research institute. We welcome the submission of suggestions for additional datasets, either using the form below or by e-mail: datacentre@scilifelab.se. All submissions will be moderated by the Portal team before they appear on the page.
Data available from research groups in Sweden
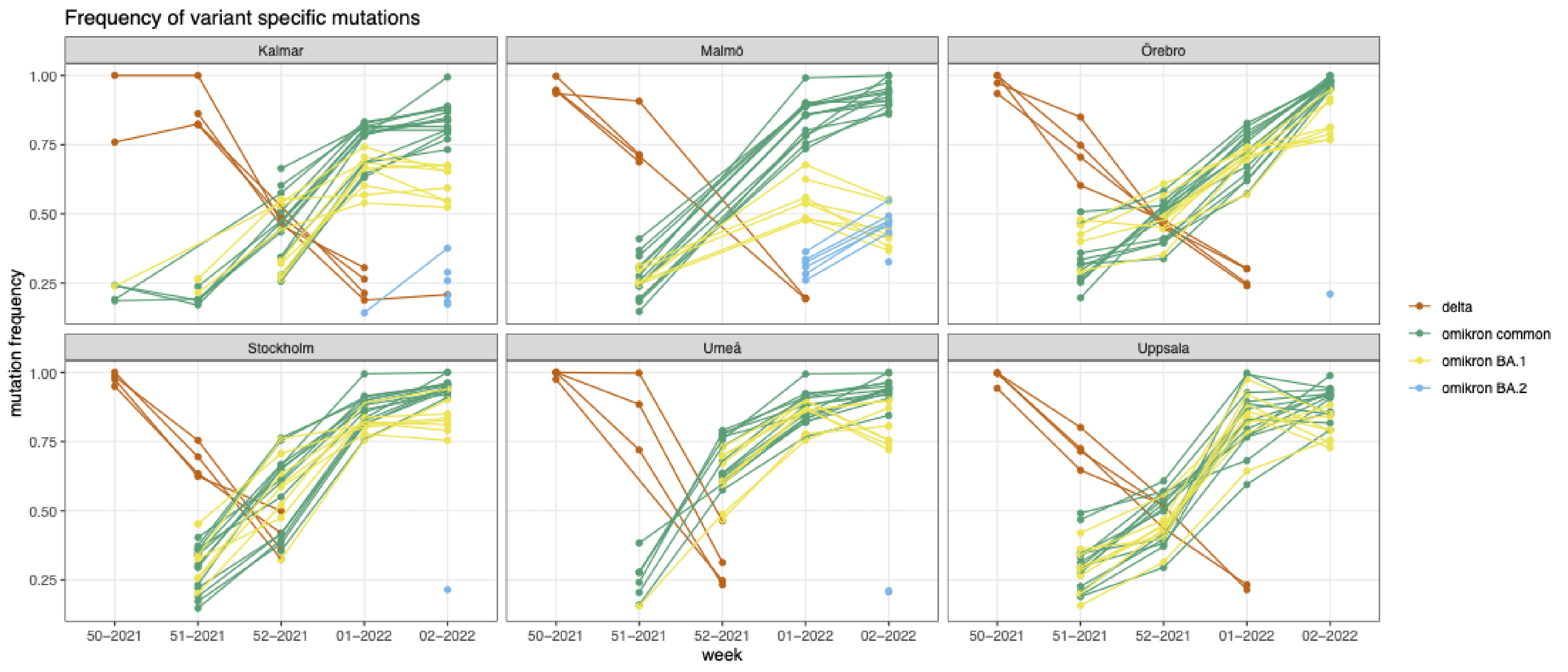
The Swedish Environmental Epidemiology Center is collecting wastewater samples from cities across Sweden and investigating frequency of variant specific SARS-CoV-2 mutations. Samples from Kalmar, Umeå, Uppsala, Örebro, Stockholm, Malmö wastewater treatment plants were analyzed. Obtained data for weeks 49-52 of 2021 and 1-2 of 2022 can be downloaded here. PIs: Maja Malmberg (maja.malmberg@slu.se), Anna J. Székely (anna.szekely@slu.se), Zeynep Çetecioğlu Gürol (zeynepcg@kth.se).
Proportion of Omicron and other variants of concern in positive samples sequenced in Region Skåne every week: Excel file for download. See also other data from Region Skåne.
Data available from research groups in other countries
From Petrillo, Querci et al.: In Silico Design of Specific Primer Sets for the Detection of B.1.1.529 SARS-CoV-2 Variant of Concern (Omicron), DOI: 10.5281/zenodo.5747871.
From Mannar, Saville et al. (DOI: 10.1101/2021.12.19.473380): Unbound Omicron spike protein trimer (PDB ID 7T9J, EMD-25759), global ACE2-bound Omicron spike protein trimer (PDB ID 7T9K, EMD-25760), focus-refinement of the ACE2-RBD interface for the ACE2-bound Omicron spike protein trimer (PDB ID 7T9L, EMD-25761).
Sequences of the Omicron variant in South Africa by the Kwazulu-Natal Research Innovation and Sequencing Platform (KRISP) and the Centre for Epidemic Response and Innovation (Viana, Moyo, et al.), BioProject PRJNA784038.
Genomic sequences of Omicron variants of SARS-CoV-2 from November 9 to November 28, 2021 (Yeh, Ting Yu; Contreras, Gregory), DOI: 10.6084/m9.figshare.17105090.v1.
Supplementary material to Structural-bioinformatics analysis of SARS-CoV-2 variants reveals higher hACE2 receptor binding affinity for Omicron B.1.1.529 spike RBD compared to wild-type reference (Durmaz, Köchl et al 2021, Research Square, DOI: 10.21203/rs.3.rs-1153124/v1): Input and final structure files as well as Pandas Dataframes of interaction, energies exported as Python Pickle files generated within this work; A GISAID acknowledgment table containing sequence data used in this study. DOI: 10.6084/m9.figshare.17129771.v1
Data on risk of SARS-CoV-2 reinfection associated with emergence of the Omicron variant in South Africa: Counts of reinfections and primary infections by province, age group (5-year bands), and sex (M, F, U) between 04 March 2020 and 27 November 2021; Daily time series of primary infections and suspected reinfections by specimen receipt date (national) (Pulliam, Juliet R.C.; van Schalkwyk, Cari; Govender, Nevashan et al.). DOI: 10.5281/zenodo.5745339.
Data Highlights
Recent data highlights related to Omicron variant. All Omicron related data highlights can be found here.
Direct RT-PCR used to monitor Omicron BA.1/BA.2 variant transition in Sweden (Update)
2022-08-15
Lentini and collegues used direct RT-PCR to show the transition from Omicron BA.1 to sub-variant BA.2 in Sweden Jan-March 2022. New Preprint from Reinius lab which shares data and code.
Ongoing research efforts
In this section, we present announcements about ongoing research efforts from researchers and organisations in Sweden. Please feel free to share what you are currently working on and/or your results, to seek collaborators to aid with aspects of your work (e.g. data, equipment, reagents, etc.). Any announcement useful for advacing research on Omicron is welcome. Feel free to submit the information either by using the form below or by sending us an e-mail to datacentre@scilifelab.se.
Omicron overcame delta in a couple weeks in analysed Swedish wastewaters
2022-01-17 17:26
By quantifying K417N and wild-type K417 mutations we estimated the ratio of omicron and delta in major cities. In all places up till Christmas delta was the dominant variant but during the holiday weeks omicron took-over. By the 10th of January delta fall below the detection limit of the assay, indicating outcompeting by omicron. For more, check out our latest press-release: https://www.slu.se/ew-nyheter/2022/1/omikron-dominerar-nu-nastan-helt-i-avloppsvattnet/
Contact: Anna Székely, anna.szekely@slu.se
Omicron VoC detected in Uppsala wastewater
2022-01-13 14:40
Anna Szekely shared first results in work attempting to detect Omicron VoC in Uppsala wastewater samples. Here they are: ‘On w51 delta dominated, omicron mutations were barely detectable. On w52 delta continued to increase but omicron also showed a strong signal (probably similar ratio of the 2 variants). On w1 delta started to decrease and omicron more than doubled.’ See Anna Szekely’s twitter account for future updates.
Contact: Arnold Kochari, arnold.kochari@scilifelab.se
Proportion of Omicron VoC in Region Skåne
2021-12-22 16:50
Region Skåne is publishing the proportion of variants in the samples sequenced every week, it can be followed on this page. During week 49, 21.8% of samples were of the Omicron VoC (out of 565 sequenced samples in total that week).
Contact: Arnold Kochari, arnold.kochari@scilifelab.se
Preliminary results on neutralization resistance of the Omicron VoC from the Murrell lab
2021-12-20 12:55
On December 7 the Murrell lab has made public two sets of prelimiary results on neutralization resistance of the Omicron VoC: Neutralization resistance of the Omicron SARS-CoV-2 Variant of Concern against monoclonal antibodies and Quantification of the neutralization resistance of the Omicron Variant of Concern. Both are prelimiary reports, subject to modification. I recommend following Ben Murrell’s Twitter account for updates.
Contact: Arnold Kochari, arnold.kochari@scilifelab.se
Exploring Omicron VoC in Sweden using Nextstrain
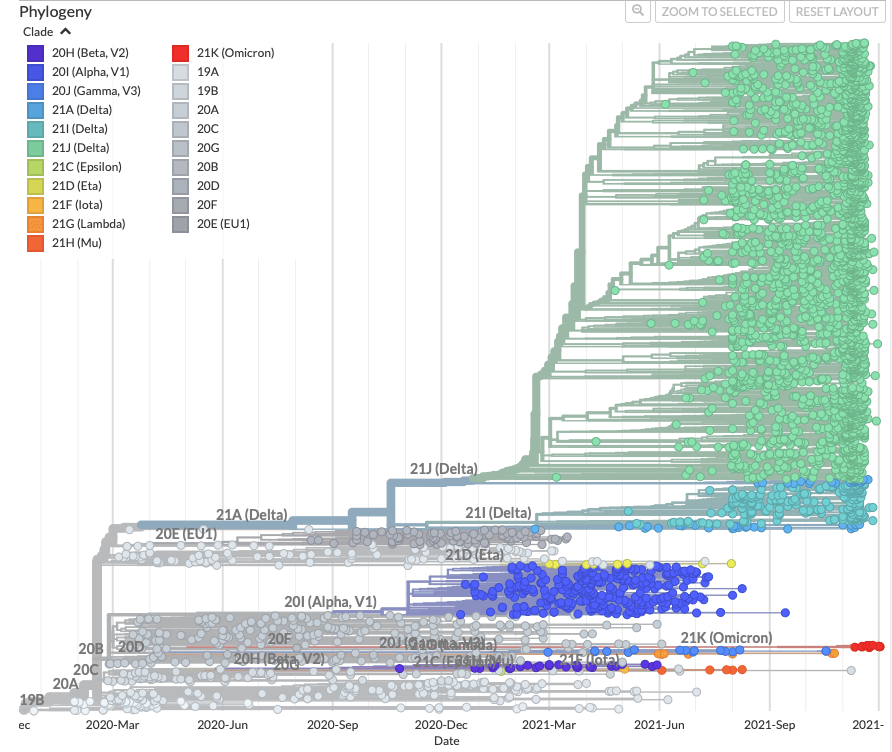
There is a Nextstrain build maintained by the Neher lab and NextStrain team that focusses specifically on sequences from Sweden, follow this link for Omicron focus. Please keep in mind that quality of the analyses by Nextstrain depends on the amount and quality of available data so the results should be taken with caution.
There are 4 main visualisation panels in Nextstrain:
- A phylogenetic tree showing the relationships between variants.
- A geographic map showing which variants have been detected in Sweden and their relative prevalence by default. Other aspects of the data related to sequences (e.g. the sampling date) can be displayed instead using the Color By dropdown menu to the left of the visualisations panel. NOTE: Some data are visible for other countries, as a sample from all over the world is needed to properly root the phylogenetic tree (see here for information on subsampling procedures).
- A ‘diversity’ plot that shows information related to differences in amino acids and nucleotides across variants.
- A frequency plot showing the relative dominance of variants.
For more information on what is shown in the panels, please see e.g. this 2018 paper by Hadfield et al., or this section on interpreting analyses in the Nextrain documentation. Further, to understand how different visuaalisations relate to each other, see the narrative written by Hodcroft et al. in 2020.
Relevant publications and preprints
Here we display a selection of preprints and journal publications with participation of research groups in Sweden reporting results on Omicron VoC.
| Publication | Published |
|---|---|
|
Genomic surveillance of omicron B.1.1.529 SARS-CoV-2 and its variants between December 2021 and March 2023 in Tamil Nadu, India-A state-wide prospective longitudinal study. Selvavinayagam ST, Suvaithenamudhan S, Yong YK, Hemashree K, Rajeshkumar M, Kumaresan A, Arthydevi P, Kannan M, Gopalan N, Vignesh R, Murugesan A, Sivasankaran MP, Sankar S, Cheedarla N, Anshad AR, Govindaraj S, Zhang Y, Tan HY, Larsson M, Saravanan S, Balakrishnan P, Kulanthaivel L, Singh K, Joseph N, Velu V, Byrareddy SN, Shankar EM, Raju S J Med Virol 96 (2) e29456. DOI: 10.1002/jmv.29456 |
2024-02-00 |
|
Impact of COVID-19 on immunocompromised populations during the Omicron era: insights from the observational population-based INFORM study. Evans RA, Dube S, Lu Y, Yates M, Arnetorp S, Barnes E, Bell S, Carty L, Evans K, Graham S, Justo N, Moss P, Venkatesan S, Yokota R, Ferreira C, McNulty R, Taylor S, Quint JK Lancet Reg Health Eur 35 100747. DOI: 10.1016/j.lanepe.2023.100747 |
2023-12-00 |
|
Effectiveness of COVID-19 Vaccines over 13 Months Covering the Period of the Emergence of the Omicron Variant in the Swedish Population. Xu Y, Li H, Kirui B, Santosa A, Gisslén M, Leach S, Wettermark B, Vanfleteren LEGW, Nyberg F Vaccines 10 (12). DOI: 10.3390/vaccines10122074 |
2022-12-05 |
|
Anti-Spike Mucosal IgA Protection against SARS-CoV-2 Omicron Infection. Havervall S, Marking U, Svensson J, Greilert-Norin N, Bacchus P, Nilsson P, Hober S, Gordon M, Blom K, Klingström J, Åberg M, Smed-Sörensen A, Thålin C N Engl J Med 387 (14) 1333-1336. DOI: 10.1056/NEJMc2209651 |
2022-10-06 |
|
Monitoring of the SARS-CoV-2 Omicron BA.1/BA.2 lineage transition in the Swedish population reveals increased viral RNA levels in BA.2 cases. Lentini A, Pereira A, Winqvist O, Reinius B Med (N Y) 3 (9) 636-643.e4. DOI: 10.1016/j.medj.2022.07.007 |
2022-09-09 |
|
Milder disease trajectory among COVID-19 patients hospitalised with the SARS-CoV-2 Omicron variant compared with the Delta variant in Norway. Stålcrantz J, Kristoffersen AB, Bøås H, Veneti L, Seppälä E, Aasand N, Hungnes O, Kvåle R, Bragstad K, Buanes EA, Whittaker R Scand J Public Health 50 (6) 676-682. DOI: 10.1177/14034948221108548 |
2022-08-00 |
|
Differences between Omicron SARS-CoV-2 RBD and other variants in their ability to interact with cell receptors and monoclonal antibodies. Giron CC, Laaksonen A, Barroso da Silva FL J Biomol Struct Dyn 1-21. DOI: 10.1080/07391102.2022.2095305 |
2022-07-09 |
|
Acute odynophagia: A new symptom of COVID-19 during the SARS-CoV-2 Omicron variant wave in Sweden. Piersiala K, Kakabas L, Bruckova A, Starkhammar M, Cardell LO J Intern Med 292 (1) 154-161. DOI: 10.1111/joim.13470 |
2022-07-00 |
|
Hospital bed occupancy for COVID-19 or with SARS-CoV-2 infection in a Swedish county during the omicron wave. Spreco A, Andersson C, Sjödahl R, Dahlström Ö, Timpka T Influenza Other Respir Viruses. DOI: 10.1111/irv.13019 |
2022-06-05 |
|
SARS-CoV-2 Omicron variants BA.1 and BA.2 both show similarly reduced disease severity of COVID-19 compared to Delta, Germany, 2021 to 2022. Sievers C, Zacher B, Ullrich A, Huska M, Fuchs S, Buda S, Haas W, Diercke M, An der Heiden M, Kröger S Euro Surveill 27 (22). DOI: 10.2807/1560-7917.ES.2022.27.22.2200396 |
2022-06-00 |
|
Neutralisation sensitivity of the SARS-CoV-2 omicron (B.1.1.529) variant: a cross-sectional study. Sheward DJ, Kim C, Ehling RA, Pankow A, Castro Dopico X, Dyrdak R, Martin DP, Reddy ST, Dillner J, Karlsson Hedestam GB, Albert J, Murrell B Lancet Infect Dis 22 (6) 813-820. DOI: 10.1016/S1473-3099(22)00129-3 |
2022-06-00 |
|
Complex Mutation Pattern of Omicron BA.2: Evading Antibodies without Losing Receptor Interactions. Kannan SR, Spratt AN, Sharma K, Goyal R, Sönnerborg A, Apparsundaram S, Lorson CL, Byrareddy SN, Singh K Int J Mol Sci 23 (10). DOI: 10.3390/ijms23105534 |
2022-05-16 |
|
Convulsions in children with COVID-19 during the Omicron wave. Ludvigsson JF Acta Paediatr 111 (5) 1023-1026. DOI: 10.1111/apa.16276 |
2022-05-00 |
|
Selection analysis identifies clusters of unusual mutational changes in Omicron lineage BA.1 that likely impact Spike function. Martin DP, Lytras S, Lucaci AG, Maier W, Grüning B, Shank SD, Weaver S, MacLean OA, Orton RJ, Lemey P, Boni MF, Tegally H, Harkins GW, Scheepers C, Bhiman JN, Everatt J, Amoako DG, San JE, Giandhari J, Sigal A, NGS-SA , Williamson C, Hsiao NY, von Gottberg A, De Klerk A, Shafer RW, Robertson DL, Wilkinson RJ, Sewell BT, Lessells R, Nekrutenko A, Greaney AJ, Starr TN, Bloom JD, Murrell B, Wilkinson E, Gupta RK, de Oliveira T, Kosakovsky Pond SL Mol Biol Evol. DOI: 10.1093/molbev/msac061 |
2022-03-24 |
|
Improved Binding Affinity of Omicron's Spike Protein for the Human Angiotensin-Converting Enzyme 2 Receptor Is the Key behind Its Increased Virulence. Kumar R, Murugan NA, Srivastava V Int J Mol Sci 23 (6). DOI: 10.3390/ijms23063409 |
2022-03-21 |
|
Human serum from SARS-CoV-2-vaccinated and COVID-19 patients shows reduced binding to the RBD of SARS-CoV-2 Omicron variant. Schubert M, Bertoglio F, Steinke S, Heine PA, Ynga-Durand MA, Maass H, Sammartino JC, Cassaniti I, Zuo F, Du L, Korn J, Milošević M, Wenzel EV, Krstanović F, Polten S, Pribanić-Matešić M, Brizić I, Baldanti F, Hammarström L, Dübel S, Šustić A, Marcotte H, Strengert M, Protić A, Piralla A, Pan-Hammarström Q, Čičin-Šain L, Hust M BMC Med 20 (1) 102. DOI: 10.1186/s12916-022-02312-5 |
2022-03-03 |
|
Risk of severe COVID-19 from the Delta and Omicron variants in relation to vaccination status, sex, age and comorbidities - surveillance results from southern Sweden, July 2021 to January 2022. Kahn F, Bonander C, Moghaddassi M, Rasmussen M, Malmqvist U, Inghammar M, Björk J Euro Surveill 27 (9). DOI: 10.2807/1560-7917.ES.2022.27.9.2200121 |
2022-03-00 |
|
Predictive performance and clinical application of COV50, a urinary proteomic biomarker in early COVID-19 infection: a cohort study Staessen JA, Wendt R, Yu YL, Thijs L, Siwy J, Raad J, Metzger J, Neuhaus B, Papkalla A, von der Leyen H, Mebazaa A, Dudoignon E, Spasovski G, Milenkova M, Canevska-Taneska A, Psichogiou M, Rajzer MW, Fulawka L, Dzitkowska-Zabielska M, Weiss G, Feldt T, Stegemann M, Normark J, Zoufaly A, Schmiedel S, Seilmaier M, Rumpf B, Banasik M, Krajewska M, Catanese L, Rupprecht H, Czerwienska B, Peters B, Nilsson A, Rothfuss K, Luebbert C, Mischak H, Beige J medRxiv. DOI: 10.1101/2022.01.20.22269599 |
2022-01-23 |
|
Novel universal SARS-CoV DNA vaccine inducing neutralizing antibodies to huCoV-19/WH01, Beta, Delta and Omicron variants and T cells to Bat-CoV Appelberg KS, Ahlen G, Nikoyan N, Yan J, Weber S, Larsson O, Höglund U, Aleman S, Weber F, Perlhamre E, Apro J, Gidlund EK, Tuvesson O, Cadossi M, Salati S, Tegel H, Hober S, Frelin L, mirazimi a, Sallberg M Research Square. DOI: 10.21203/rs.3.rs-1276351/v1 |
2022-01-20 |
|
Selection analysis identifies unusual clustered mutational changes in Omicron lineage BA.1 that likely impact Spike function. Martin DP, Lytras S, Lucaci AG, Maier W, Grüning B, Shank SD, Weaver S, MacLean OA, Orton RJ, Lemey P, Boni MF, Tegally H, Harkins G, Scheepers C, Bhiman JN, Everatt J, Amoako DG, San JE, Giandhari J, Sigal A, NGS-SA , Williamson C, Hsiao NY, von Gottberg A, De Klerk A, Shafer RW, Robertson DL, Wilkinson RJ, Sewell BT, Lessells R, Nekrutenko A, Greaney AJ, Starr TN, Bloom JD, Murrell B, Wilkinson E, Gupta RK, de Oliveira T, Kosakovsky Pond SL bioRxiv. DOI: 10.1101/2022.01.14.476382 |
2022-01-18 |
|
Towards an optimal monoclonal antibody with higher binding affinity to the receptor-binding domain of SARS-CoV-2 spike proteins from different variants Neamtu A, Mocci F, Laaksonen A, Barroso da Silva FL bioRxiv. DOI: 10.1101/2022.01.04.474958 |
2022-01-05 |
|
Heterologous immunization with inactivated vaccine followed by mRNA booster elicits strong humoral and cellular immune responses against the SARS-CoV-2 Omicron variant Zuo F, Abolhassani H, Du L, Piralla A, Bertoglio F, de Campos-Mata L, Wan H, Schubert M, Wang Y, Sun R, Cassaniti I, Vlachiotis S, Kumagai-Braesch M, Andréll J, Zhang Z, Xue Y, Wenzel EV, Calzolai L, Varani L, Rezaei N, Chavoshzadeh Z, Baldanti F, Hust M, Hammarström L, Marcotte H, Pan-Hammarström Q medRxiv. DOI: 10.1101/2022.01.04.22268755 |
2022-01-05 |
|
Structural basis of Omicron neutralization by affinity-matured public antibodies Sheward DJ, Pushparaj P, Das H, Kim C, Kim S, Hanke L, Dyrdak R, McInerney G, Albert J, Murrell B, Karlsson Hedestam GB, Hällberg BM bioRxiv. DOI: 10.1101/2022.01.03.474825 |
2022-01-04 |
|
Reduced risk of hospitalisation among reported COVID-19 cases infected with the SARS-CoV-2 Omicron BA.1 variant compared with the Delta variant, Norway, December 2021 to January 2022. Veneti L, Bøås H, Bråthen Kristoffersen A, Stålcrantz J, Bragstad K, Hungnes O, Storm ML, Aasand N, Rø G, Starrfelt J, Seppälä E, Kvåle R, Vold L, Nygård K, Buanes EA, Whittaker R Euro Surveill 27 (4). DOI: 10.2807/1560-7917.ES.2022.27.4.2200077 |
2022-01-00 |
|
Influenza A H1N1–mediated pre-existing immunity to SARS-CoV-2 predicts COVID-19 outbreak dynamics Almazán NM, Rahbar A, Carlsson M, Hoffman T, Kolstad L, Rönnberg B, Pantalone MR, Fuchs IL, Nauclér A, Ohlin M, Sacharczuk M, Religa P, Amér S, Molnár C, Lundkvist Å, Susrud A, Sörensen B, Söderberg-Nauclér C medRxiv. DOI: 10.1101/2021.12.23.21268321 |
2021-12-25 |
|
Omicron SARS-CoV-2 variant: Unique features and their impact on pre-existing antibodies. Kannan SR, Spratt AN, Sharma K, Chand HS, Byrareddy SN, Singh K J Autoimmun 126 102779. DOI: 10.1016/j.jaut.2021.102779 |
2021-12-13 |
Other useful resources
Here we list a number of useful resources related to the new SARS-CoV-2 Omicron variant.
- Information from the WHO about Omicon as a variant of concern (2021-11-26)
- Epidemiological reports from ECDC
- Data on SARS-CoV-2 variants in the EU/EEA from ECDC
- Information about Omicron from the CDC
- Information about Omicron from the Statens Serum Institute, Denmark
- Statens Serum Institute, Denmark Rapport Dec 12 2021
- Statistics about cases of Omicron from the Danish Statens Serum Institute
- Information about Omicron from the Norwegian Institute of Public Health
- For lineages and information related to sequences from Cov-lineages
- Lineages and information related to sequences from Nextstrain
- Assembled information about the Omicron from the PHA4GE (Bioinformatics Pipelines & Visualization Working Group)
- VEO report on mutations and variation in publicly shared SARS-CoV-2 raw sequencing data, Report No. 10, 15 December 2021
- Proportion of different variants of concern in positive samples sequenced in Region Skåne
- Ontario COVID-19 Dashboard: Tracking Omicron
- ESCMID Urgent update on Omicron SARS-CoV-2 variant, 2021-12-22
- Statens Serum Institut: Antigen tests can detect new variants, 2022-01-07
- Nextstrain: Phylogenetic analysis of SARS-CoV-2 clusters in their international context - cluster DanishCluster
- WHO Statement on Omicron sublineage BA.2
Get in touch with us
Do you have suggestions or other feedback? Get in touch with us by e-mailing datacentre@scilifelab.se.