It is necessary to do some preparation of files before submission, regardless of the eventual submission route that you select. This page explains how to perform these initial steps, so that you can successfully complete your submission.
Required file formats
All sequence files that are submitted to ENA must be in an appropriate file format to be accepted. Details of the formats are listed in the Terminology and Metadata tab. Once all the files are in order, you can start the submission process.
Obtaining an ENA Webin Account
In order to do a submission, you need an account in ENA. To create an account, please go to the ENA submission homepage, click on Register, and fill in the required details.
Login page on the ENA submission homepage:
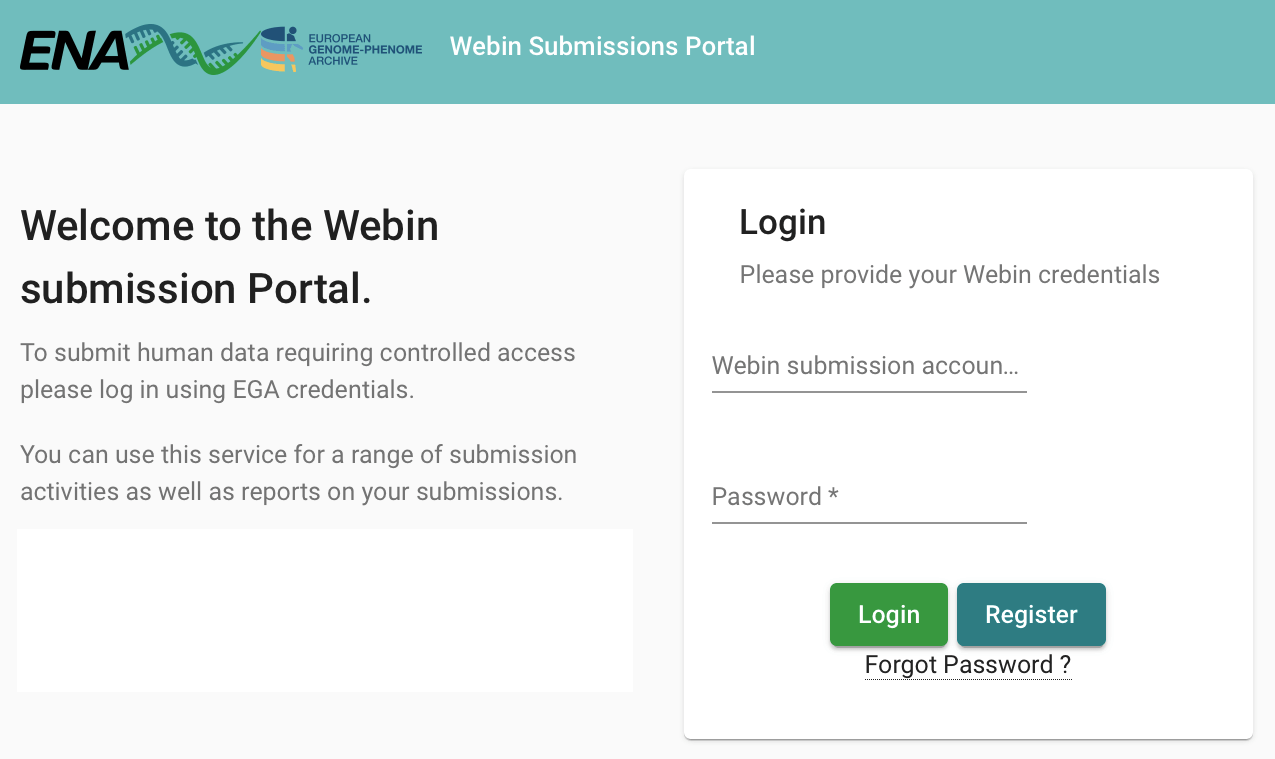
Enter account details to obtain an account in ENA:
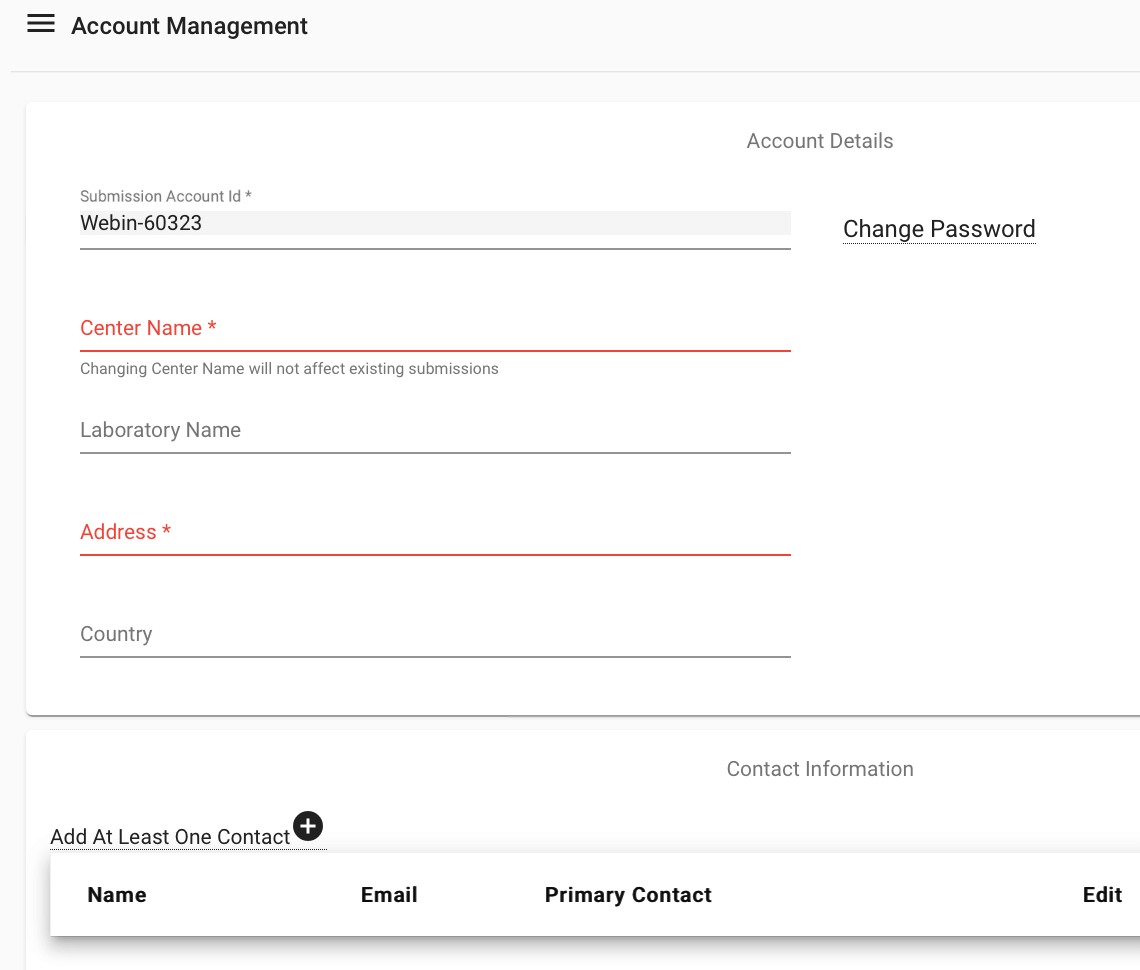
Once you have filled in all of the required fields, you can log in to the submission service. You may wish to add contacts if you are working with other people to complete your submission. You can do this by logging into your account then selecting Home > Manage account. Any contacts that you add will be notified if there are any major changes to data submissions, and they will be listed as contacts on public data.
Obtaining example data
You are welcome to follow this tutorial using your own data. However, you can instead use example data, which you can download as a single zip file. Using the example can be helpful for familiarising yourself with how to structure your data for submission and the steps required to complete a submission.
The example data was originally produced by ENA, but we have restructured it for use with the two submission routes described in this tutorial. With the example data, you can submit 3 different samples to an example project, together with raw read data and sequences associated with each sample.
Whether you use your own data or the example data, we recommend that you write the following code in a terminal window to create a path variable called WORKSHOP (this will save you from having to type the filepath repeatedly):
export WORKSHOP="/path/to/example_data/"
cd $WORKSHOP/
ls
Preparing the metadata
To ensure that sample data is registered with at least a minimum amount of metadata, ENA provides “Sample Checklists” which are used during registration to tailor the sample descriptions to fit minimum standards. The most appropriate checklist for SARS-CoV-2 viral submissions is the ENA virus pathogen reporting standard checklist (ERC000033). This includes 9 mandatory, 15 recommended, and 11 optional fields (along with additional user-defined fields that can be used). Please note that some fields are free text, while others have controlled vocabulary.
In order to ensure that your SARS-CoV-2 data is properly described, we recommend downloading and filling in this metadata template (Excel). This template contains not only the ENA checklist for the data, but also describes all levels of metadata required for a submission. The template is divided into five sheets (each related to a type of metadata object), namely; study, sample, experiment, run and assemblies. The experiment and run sheets are used for describing the raw reads, and the assemblies sheet is used for describing the sequence assembly. It is good practice to fill in all relevant sheets in the template, as having all the metadata collected in one place eases the submission process.
In the sample sheet, the first row is the ENA virus sample checklist field, the second row is the ENA definition (provides a description of the field), and the third row is ENA requirement status (whether the field is mandatory, recommended or optional). When you populate the sheet with your metadata, you can delete the second and third row. The default values for SARS-CoV2 submissions are pre-filled (in red) for relevant fields. Some fields have controlled vocabulary, which are available in the template as drop-down lists (the lists become visible when you click on a cell).
For your convenience, we also provide this pre-filled version of the metadata template (Excel), so that you can see how the template should be populated for use with the example data. This can also help with understanding how to fill in such a template for your own data.
Note: It is strongly recommended that you provide as much information as possible in the metadata sheets. This will increase the FAIRness of your data, and thus the probability that it will be useful in future research efforts.
The sample metadata sheet can be submitted to ENA, however it must be in .tsv file format. For ease, the downloads on this page are provided in .xlsx, commonly known as Microsoft Excel, format. It is therefore necessary for you to convert the file format. If you are using Microsoft Excel to edit the metadata templates, click File (or the Office Button) > Save As and change ‘save as file type’ to ‘Text (Tab delimited) (*. tsv)’. Enter a file name and save the file. If you are using a Mac, you can change the file type by right-clicking on the .xlsx file in Finder, selecting rename and typing ‘.tsv’ at the end of your file name.