Host factor interaction between human G3BP proteins and SARS-CoV-2 N-protein could represent an antiviral target
Published: 2021-11-24
The current COVID-19 pandemic has challenged societies world-wide over the last two years. Although vaccines have been developed in record time, and therapeutic drugs have been developed and repurposed, the pandemic is still challenging healthcare. More treatment options (e.g. novel antivirals) are warranted in preparation for future pandemic outbreaks.
RNA viruses, such as coronaviruses, ebola, and dengue, can cause a variety of diseases and pose a threat to public health. The genomic RNA of coronaviruses encode around 30 viral proteins. These proteins exhibit unique functions and can interact with host cellular proteins; allowing viral proliferation and potential immune escape. Disruption of the virus-host factor interactions has been proposed as a mechanism of novel antiviral reagents. The rationale behind this is that, by targeting interactions known to be used by multiple viruses, the antivirals would have broader spectrum activity.
In a recently published article in Nature Communications, researchers from Sweden, Denmark and the UK (first authors Thomas Kruse (University of Copenhagen) and Caroline Benz (Uppsala University), and corresponding authors Prof. Ylva Ivarsson (Uppsala University), Prof. Jakob Nilsson (University of Copenhagen), and Prof. Anna K Överby (Umeå University)) describe a scalable viral peptide discovery approach covering 229 RNA viruses. This approach provides high resolution information on direct virus-host interactions.
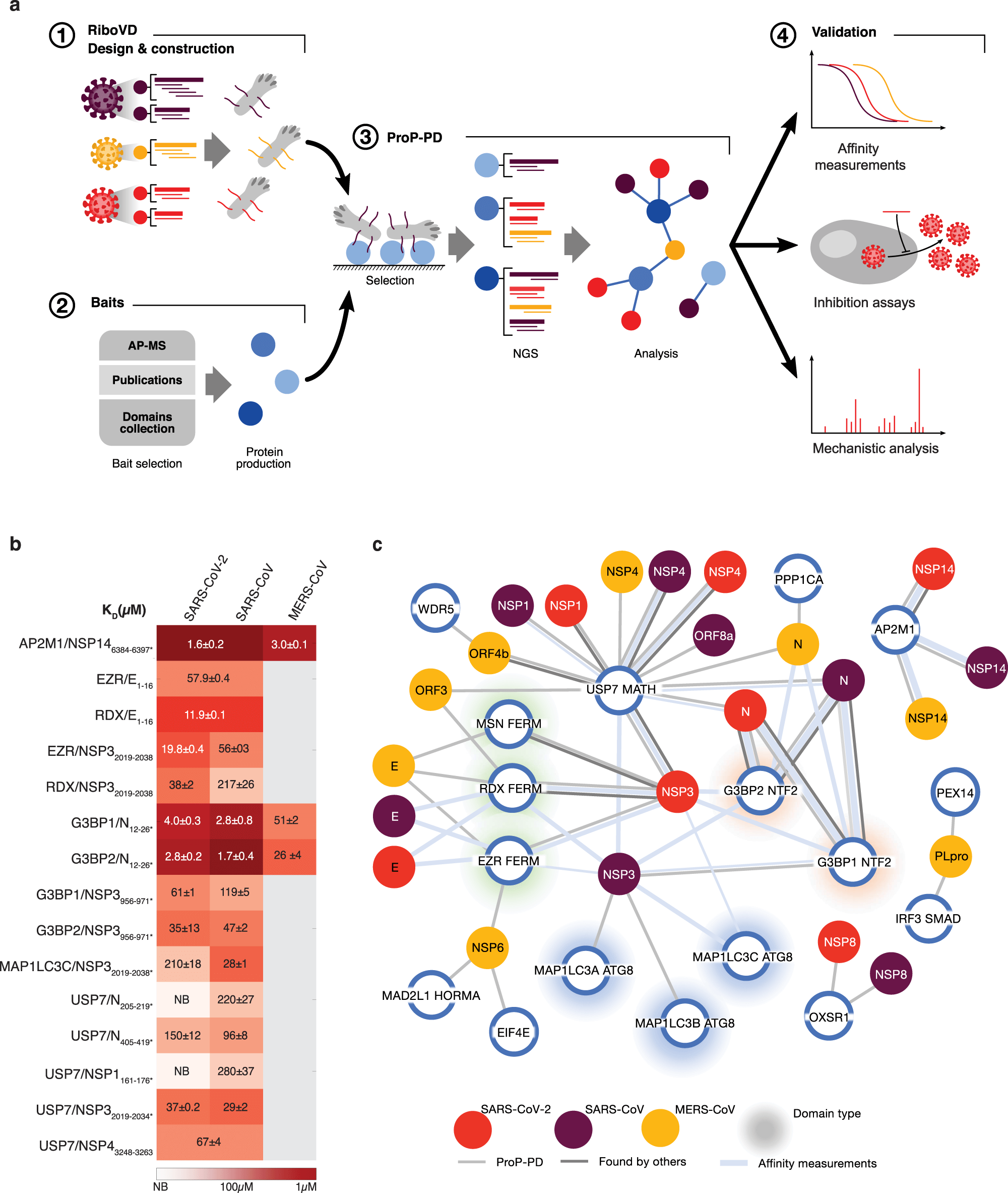
Viral proteins often use short linear interaction motifs (SLiMs) found in unstructured regions of proteins to “hijack” cellular host factors. Previous research has shown that the ProP-PD (Proteomic peptide-phage display) technology can be used to identify SLiMs binding to host factors and identify novel SLiM-based interactions and binding sites.
Therefore, Kruse, Benz, and colleagues prepared a unique phage display library (RiboVD library). The library contains unstructured regions of viral proteins from >200 RNA viruses, including SARS-CoV-2 as well as other related viruses like SARS-CoV, MERS-CoV, and 19 additional coronavirus strains. Although this study focuses on coronaviruses, the library could be used for any RNA virus.
Using the viral peptide discovery approach, Kruse and Benz et al. found 269 putative SLiM-based interactions for 18 coronaviruses. The researchers found that almost half (43%) of the interaction pairs involved human coronavirus proteins. They then validated a number of interactions using fluorescence polarisation (FP) affinity (N = 27) and visualised information for both human and bat coronavirus proteins in an extensive network. In addition, the researchers also prepared a map of the viral proteins mediating SARS-CoV-2, SARS-CoV, and MERS-CoV interactions with human host factors. Some unique interaction properties for these three coronaviruses were identified as a result, and a specific interaction between the human G3BP proteins and SARS-CoV-2 nucleocapsid (N) protein was found. This has important implications, as the SARS-CoV-2 N-protein is involved in viral replication and viral packaging. The N-G3BP interaction is therefore a potential target for novel antiviral drugs.
In order to further study the ability of the N-protein to inhibit stress granule formation, the researchers first purified G3BP1-YFP from HeLa cells and then performed quantitative label free mass spectrometry to determine the proteins being specifically displaced by the N wt peptide. This showed that the N-protein displaced 59 proteins, including several nuclear pore complex components. The results from the mass spectrometry data can be used for future studies of both basic stress granule biology and G3BP signalling.
In summary, Kruse, Benz, and colleagues showed that the viral peptide discovery approach could identify novel specific SLiM mediated host factor interactions for SARS-CoV-2 that can provide mechanistic insights for antiviral intervention. In addition, the approach can also be used to study a number of other RNA viruses, like ebola and dengue, that have the potential to cause future pandemics.
“Our study gives high-resolution information on the interactions that SARS-CoV-2, and other viruses, use to take over the host cell. The novel database of SLiM-based host-pathogen interactions we build up and share with the community will contribute to our preparedness for coming pandemics.“ says Ylva Ivarsson, Professor at Uppsala University.
The article was published open access online in Nature Communications on 19th November 2021 (Accepted 6th October 2021) under licence CC BY 4.O. The authors shared the first pre-print on bioRxiv on 19th April 2021.
The researchers have shared mass spectrometry proteomics data in the ProteomeXchange Consortium via the PRIDE partner repository, with the dataset identifier PXD025410. The results from the protein-protein interactions will be deposited in the IntAct Molecular Interaction Database. Supplementary material is also available in the full text version of the article here.
This work has received grants from the Swedish Foundation for Strategic Research, the Swedish Research Council, the Knut and Alice Wallenberg Foundation, Sygeforsikering Denmark, Cancer Research UK, and the Novo Nordisk Foundation.
For more information see the press release from Uppsala University here.
Data
- Mass spectrometry proteomics data are available in the ProteomeXchange Consortium via the PRIDE partner (dataset identifier: PXD025410)
- Protein-protein interactions will be deposited in the IntAct Molecular Interaction Database
- A complete list of viral strains in the RiboVD library
- Supplementary material is also available in the full text version of the article
Article
DOI: 10.1038/s41467-021-26498-z
Kruse, T., Benz, C., Garvanska, D. H., Lindqvist, R., Mihalic, F., Coscia, F., Inturi, R., Sayadi, A., Simonetti, L., Nilsson, E., Ali, M., Kliche, J., Morro, A. M., Mund, A., Andersson, E., McInerney, G., Mann, M., Jemth, P., Davey, N. E., Överby, A. K., Nilsson J. & Ivarsson, Y. Large scale discovery of coronavirus-host factor protein interaction motifs reveals SARS-CoV-2 specific mechanisms and vulnerabilities. Nature Communications 12, 6761 (2021).