SARS-CoV-2 Reporter for visualisation of SARS-CoV-2 genome sequence data (SC2reporter)
Funding given for project project entitled:
Genomic Pandemic Preparedness Portfolio (G3P) (PLP1 capability)
PI(s)/Head responsible for the resource:
Jonas Björkman (Region Skåne)
Host organisation(s):
Multiple organisations contribute to SC2reporter; Region Skåne (initial developer), Lund University, Karolinska Institutet, Karolinska University Hospital, GMS
Resource description:
A web-based visualisation application for the ongoing SARS-CoV-2 pandemic. The tool is primarily used for epidemiological analysis where pipeline analysis results and metadata are loaded into a mongoDB database. The SNVs and indels can then be compared between all isolated in the database.
For more information on the Pandemic Laboratory Preparedness resources associated with this subproject, see Genomic Pandemic Preparedness Portfolio (G3P). Please also refer to other associated subprojects; GMS Arctic and taxprofiler.
Below is a still image of one of the visualisations produced using SC2reporter.
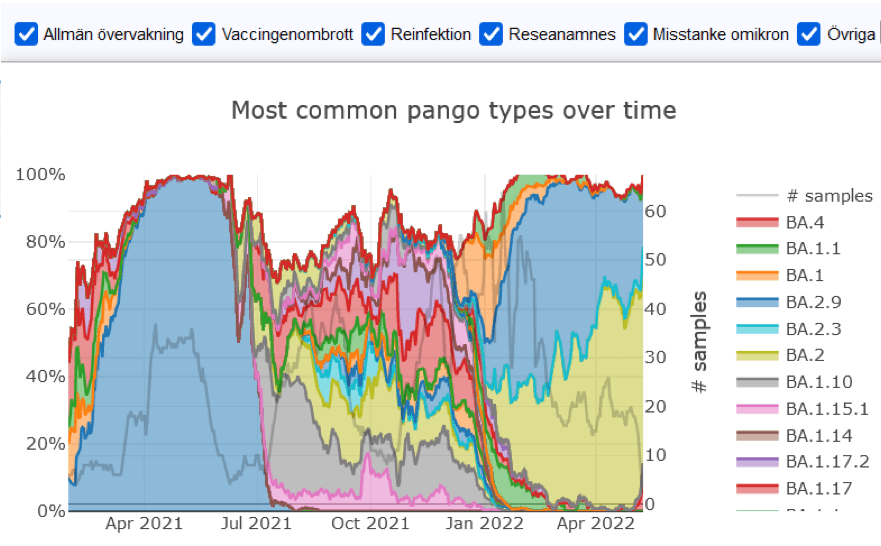
How this resource can be used for Pandemic Preparedness research:
This resource can be used to explore SARS-CoV-2 sequence data. The tool has multiple features:
- List all variants a single isolate contains. This includes nuc and aa changes over the complete genome including the coverage, frequency, and prevalence of the mutation in your own dataset.
- Compare the isolate with all other isolates in your database within a set number of variant differences.
- Show pango and nextstrain clades
- Every mutation and type is hyperlinked to external resources for additional information
- A dashboard that gives an overview of types over time
- A rerun tool that helps evaluate reruns of isolates in the lab.
- Visualise phylogenetic trees of selected isolates
We are currently working on a new release where we will be:
Who and under which conditions is able to access the resource:
SC2Reporter is available free-of-charge on GitHub.
Available data, code, and protocols from the resource:
All code related to SC2Reporter is available on GitHub.
SOPs, guidelines, publications, etc. describing the resource:
Instructions about how to use SC2Reporter and relevant reports are available on GitHub.
Webpage:
https://github.com/genomic-medicine-sweden/sc2reporter
Contact information:
Jonas Björkman
Molecular Biologist
Email: Jonas.bjorkman@skane.se